Instantaneous Brain-to-Voice Neuroprosthesis
A remarkable new study presents the first brain-to-voice neuroprosthesis that synthesizes speech directly from intracortical neural activity in real time.
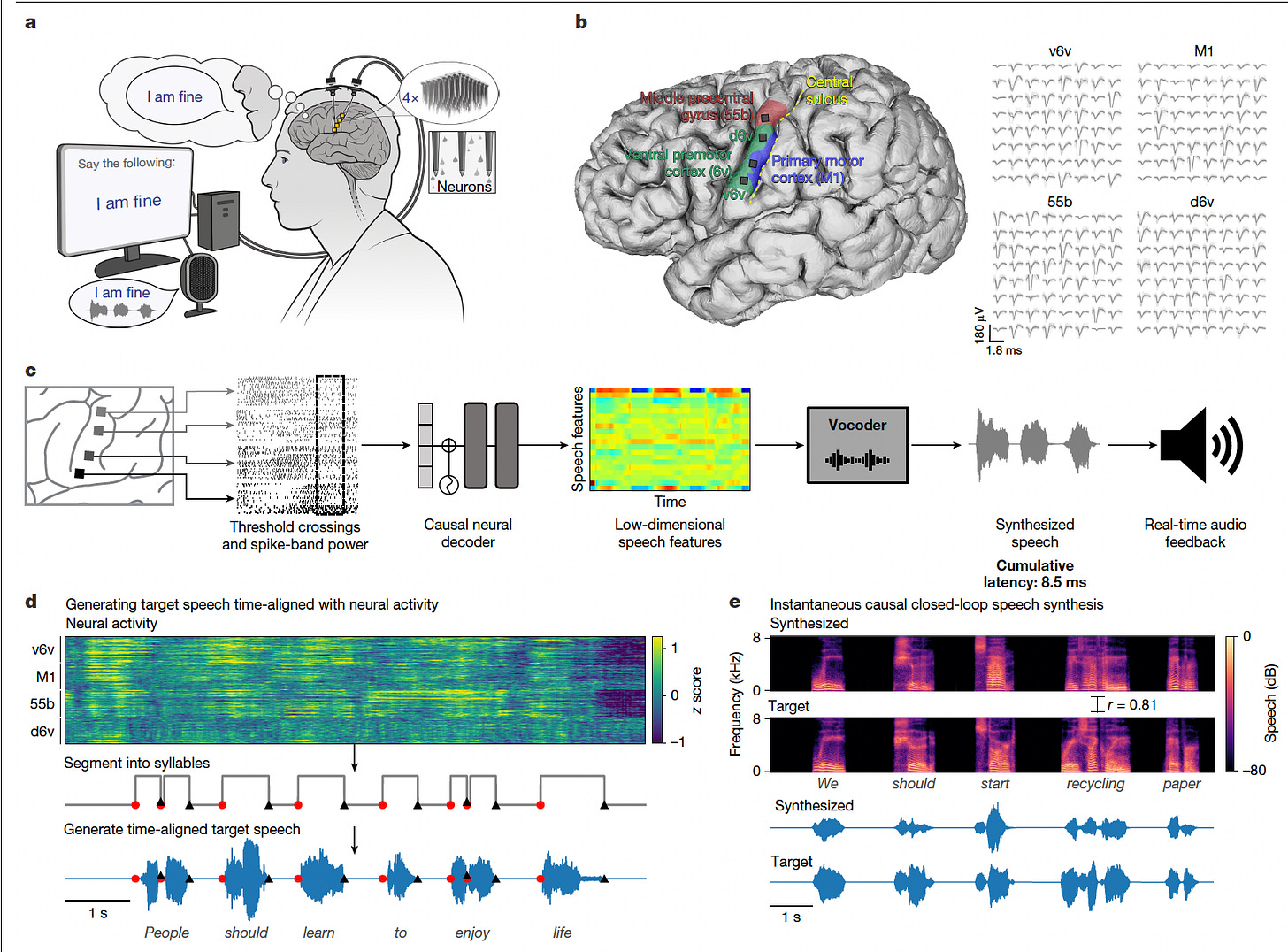
Researchers implanted 256 microelectrodes in the motor cortex of a man with ALS and severe dysarthria, enabling him to generate an audible, intelligible voice as he attempted to speak. The system provided immediate audio feedback, capturing both the content and expressive qualities of his speech.
Trained a Transformer-based neural decoder to synthesize speech features from intracortical signals with just 10 ms of causal neural data, enabling real-time voice output without relying on a limited vocabulary or predefined speech tokens.
Overcame the absence of ground-truth speech data by time-aligning text-to-speech-generated waveforms with neural activity, allowing accurate training even when the participant’s own voice was unintelligible.
Achieved strong intelligibility: listeners correctly matched synthesized sentences with transcripts 94.3% of the time, and word error rates dropped from 96.4% (residual speech) to 43.8% using the BCI.
Enabled expressive control over voice output, allowing the participant to change intonation for questions, emphasize specific words, sing melodies across pitch levels, and even replicate a personalized version of his pre-ALS voice.
This work demonstrates a transformative leap in restoring spoken communication, merging speech content with expressive voice features through a fully closed-loop brain-computer interface.
RNAGenesis: A Generalist Foundation Model for Functional RNA Therapeutics
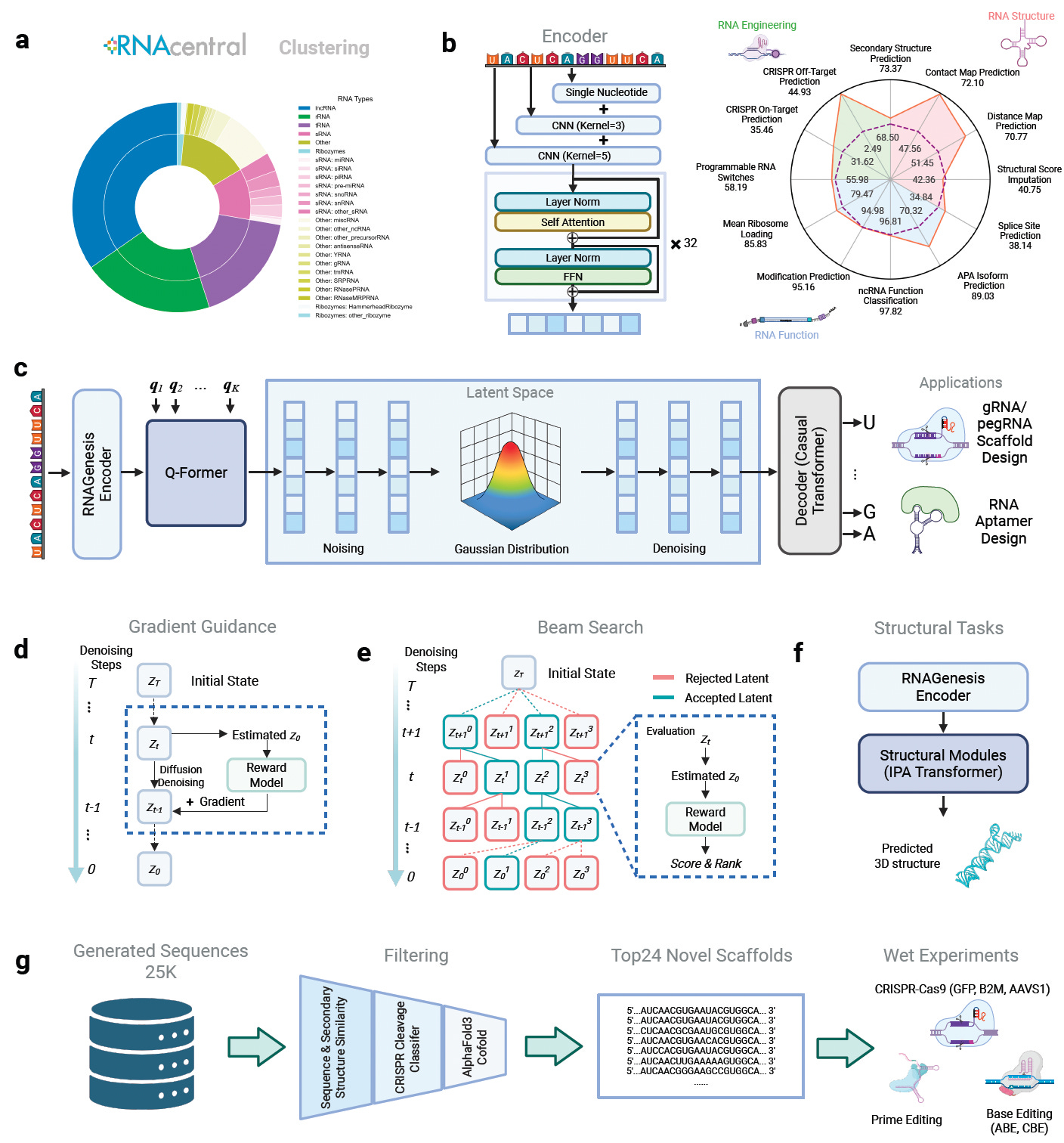
Stanford and Princeton researchers introduce RNAGenesis, a foundation model for RNA that unifies sequence understanding, structure prediction, and de novo therapeutic design.
Unlike prior RNA models focused solely on representation learning, RNAGenesis integrates a latent diffusion framework with structural inference to design aptamers, sgRNAs, and other functional RNAs. It achieves state-of-the-art performance across predictive and generative RNA tasks, validated both computationally and in wet-lab experiments.
Surpassed previous models on 11 out of 13 BEACON benchmark tasks, achieving high accuracy in structure prediction, non-coding RNA classification, and regulatory function modeling.
Outperformed RNA-FM and Evo2 in predicting efficacy for RNA therapeutics including ASOs, siRNAs, shRNAs, circRNAs, and UTR variants using the newly introduced RNATx-Bench dataset with over 100,000 labeled sequences.
Enabled de novo aptamer and sgRNA scaffold design through latent diffusion and inference-time alignment, generating RNA sequences with improved thermodynamic stability, binding affinity (KD as low as 4.02 nM), and CRISPR editing efficiency (up to 2.5x improvement).
Demonstrated strong zero-shot generalization across genome editing systems including CRISPR-Cas9, base editing, and prime editing, with designed scaffolds maintaining native-like structures and forming more hydrogen bonds with Cas9.
RNAGenesis establishes a unified and generalist RNA foundation model with broad applicability across therapeutic RNA design, structural biology, and precision genome engineering.
ChatNT: A Conversational Agent for Genomics Tasks
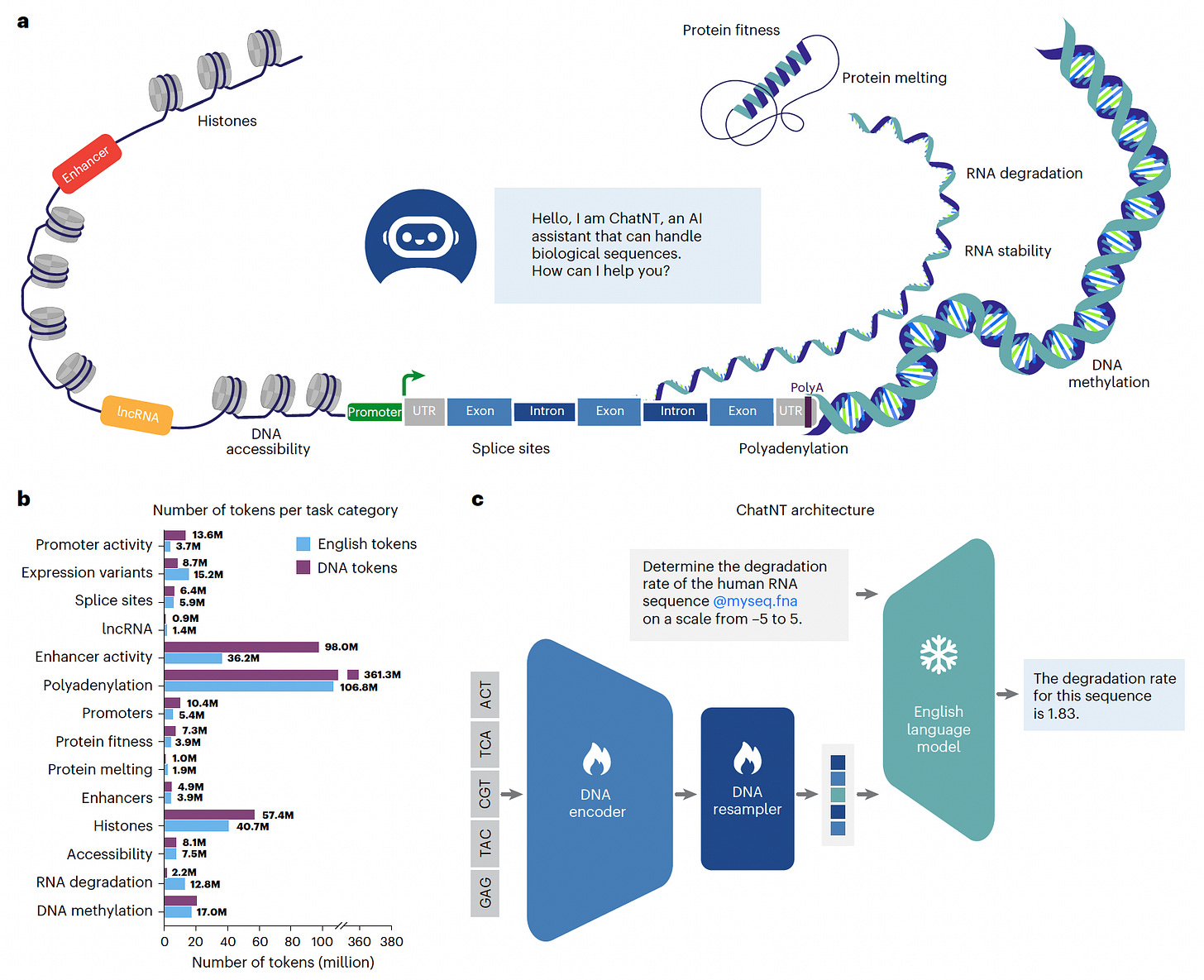
A new study introduces ChatNT, a multimodal conversational agent capable of solving biologically relevant tasks involving DNA, RNA, and protein sequences.
Built by combining a DNA foundation model with an instruction-tuned English decoder, ChatNT enables users to query biological sequences in natural language. It achieves state-of-the-art performance while being accessible to users with no programming background.
Outperformed all prior models on the Nucleotide Transformer benchmark, achieving a mean Matthews correlation coefficient (MCC) of 0.77 across 18 genomics tasks, improving over previous bests by 8 points.
Framed 27 genomics tasks, including classification and regression, as English-language instructions spanning processes like methylation, splicing, polyadenylation, and protein melting point prediction.
Matched or exceeded specialized models such as APARENT2 and ESM2 on tasks like RNA polyadenylation (PCC: 0.91) and protein melting point prediction (PCC: 0.89), all within a single unified model.
Used attribution and calibration techniques to interpret predictions and assess confidence, identifying biologically meaningful features like TATA motifs and splice site dinucleotides.
ChatNT marks a pivotal shift toward general-purpose biological agents by merging genomic understanding with the accessibility of language-based interaction.
COIN: Annotation-Free Cell Segmentation with Confidence-Guided Distillation
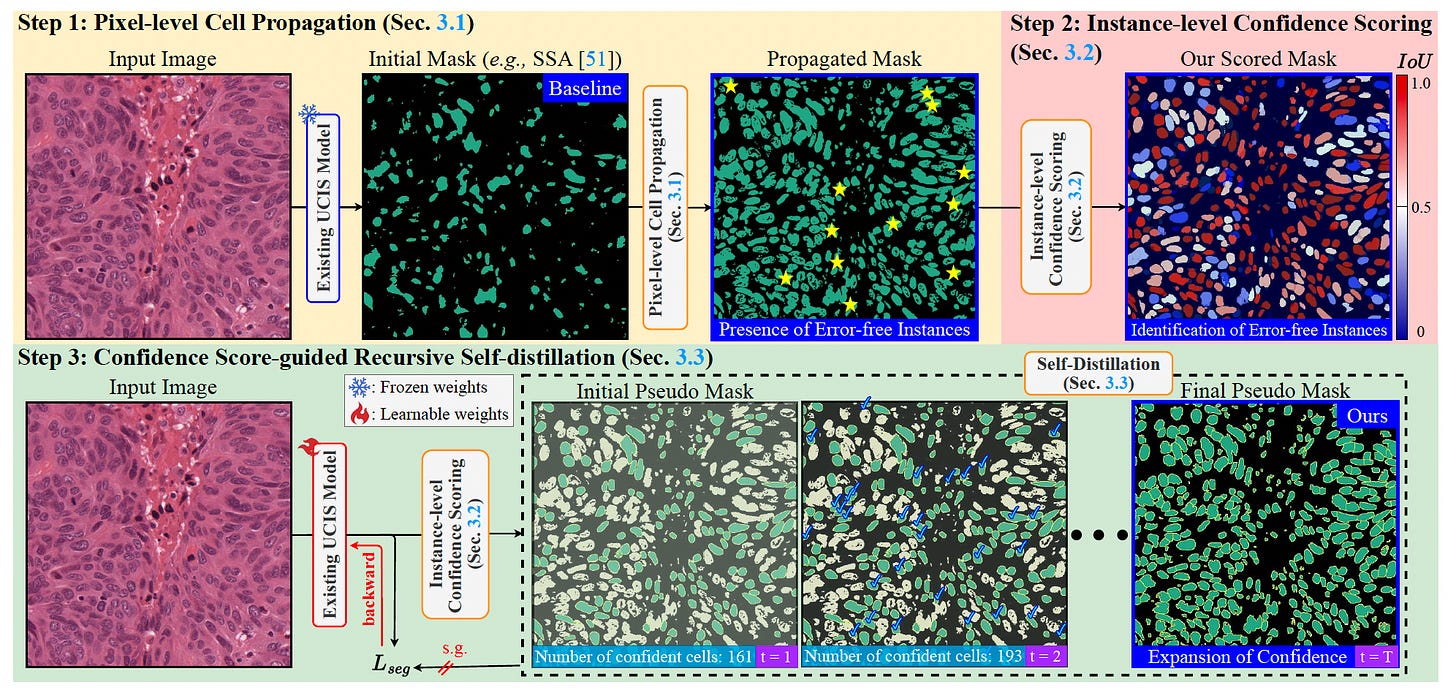
A new framework, COIN, tackles the critical challenge in unsupervised cell instance segmentation (UCIS): the complete absence of error-free instances. Compared to previous UCIS methods that struggle to delineate individual cell boundaries, COIN introduces a three-step distillation process to extract and refine high-confidence cell instances without any manual annotations.
This innovation enables significantly improved segmentation accuracy while remaining fully annotation-free.
COIN not only outperforms all existing UCIS models but also surpasses semi- and weakly-supervised approaches on key benchmarks like MoNuSeg and TNBC, achieving over +32 percentage point improvement in instance-level performance.
Began with pixel-level propagation using unsupervised semantic segmentation and refined it using optimal transport, drastically reducing false negatives and improving cell boundary definition.
Scored each proposed instance for confidence by comparing model outputs with pseudo-labels from SAM, filtering out uncertain or erroneous instances to simulate supervision.
Applied recursive self-distillation using only the most confident instances, progressively expanding the set of high-quality training examples and enhancing model generalizability.
Achieved state-of-the-art results on six datasets including MoNuSeg, TNBC, BRCA, and PanNuke, doubling the AJI performance of baselines and even outperforming methods that rely on point or box annotations.
By eliminating the need for any cell-level annotations and still improving segmentation performance, COIN presents a major step forward for scalable and accessible histopathology image analysis.
WBM: A Foundation Model for Behavioral Data from Wearables
A new study from Apple researchers presents WBM, a foundation model trained on behavioral data from wearables to improve health prediction across both static and dynamic health states.
WBM leverages over 2.5 billion hours of aggregated behavioral data from 162,000 individuals, capturing patterns such as physical activity, mobility, and cardiovascular fitness. By optimizing tokenization and model architecture specifically for irregular time series data, the model sets a new benchmark for health monitoring using wearable-derived signals.
Trained a large-scale encoder-only foundation model on 15 million weeks of behavioral wearable data, using a simple Time Series Transformer tokenizer and a Mamba-2 backbone optimized for irregular sampling.
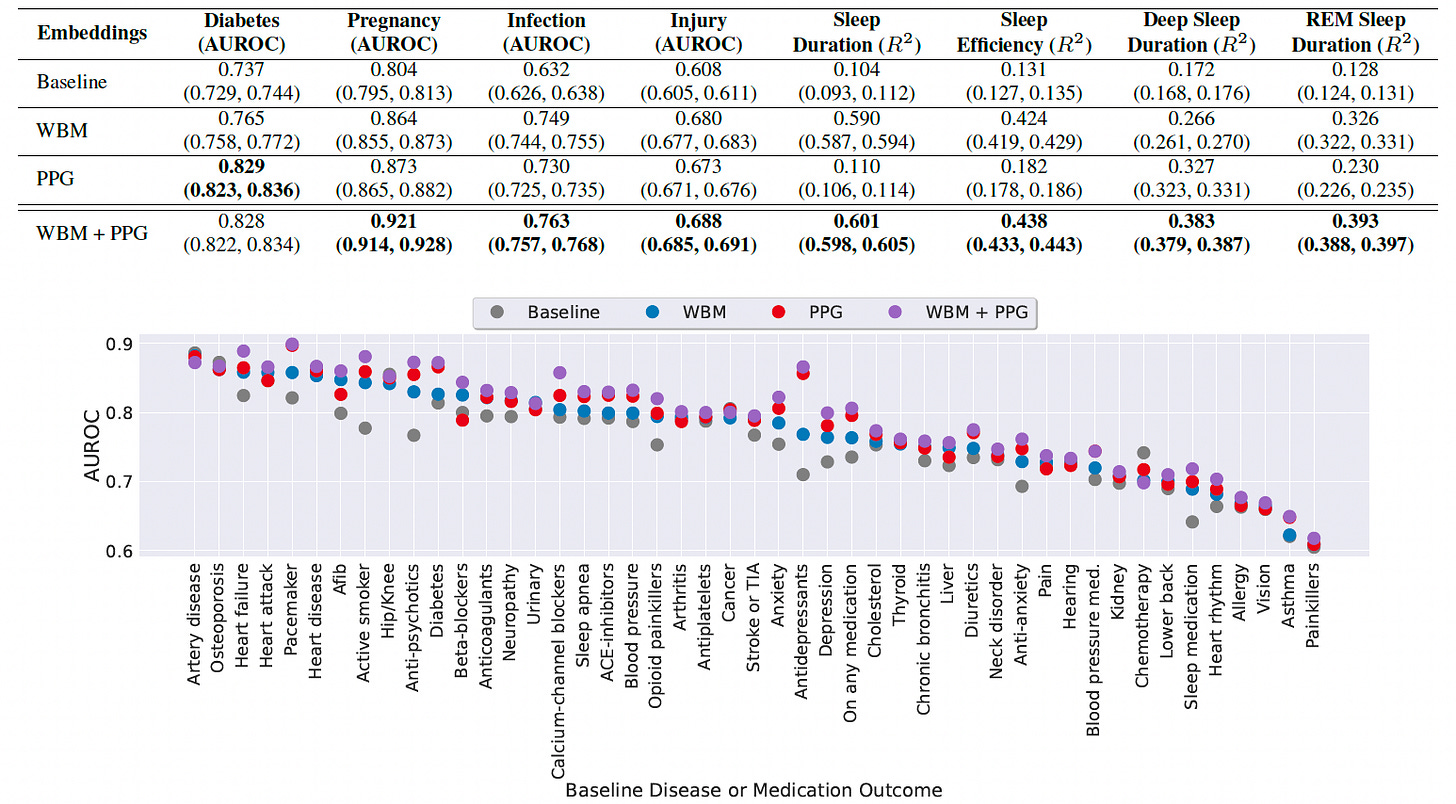
Demonstrated strong performance on 57 downstream health tasks, including static conditions (e.g., hypertension history) and dynamic states (e.g., weekly sleep metrics, respiratory infections), significantly outperforming baselines.
Outperformed models based solely on raw sensor data for behavior-driven tasks like pregnancy detection (AUROC: 0.864) and sleep prediction (R² up to 0.59), with further gains when combined with PPG embeddings.
Validated that combining behavioral and physiological data improves prediction in 42 of 47 baseline health outcomes, showing complementary value for personalized, real-world health detection.
WBM shifts the focus of wearable-based AI from raw biosignals to human-centered behavioral patterns, offering a new foundation for digital health applications in monitoring, diagnosis, and prevention.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun