Google's TxGemma, VLM for 3D Med Imaging, Text-Guided Protein Design 🚀
Health Intelligence (HINT)
2025-03-31
🚀
New Developments in Research
TxGemma: Efficient and Agentic LLMs for Therapeutics
Researchers at Google introduce TxGemma, a suite of efficient, generalist large language models (LLMs) designed for therapeutic property prediction, interactive reasoning, and explainability. Unlike task-specific models, TxGemma synthesizes information from diverse sources, facilitating broad applications across the therapeutic development pipeline.
TxGemma outperforms state-of-the-art generalist and specialist models across 66 therapeutic development tasks, offering a scalable alternative for AI-driven drug discovery.
Trained on a comprehensive dataset encompassing small molecules, proteins, nucleic acids, diseases, and cell lines, spanning 2B, 9B, and 27B parameter models.
Demonstrated superior performance over leading generalist models on 64 tasks and over specialist models on 50, highlighting its versatility in predictive and generative tasks.
Enabled efficient fine-tuning for clinical trial adverse event prediction, requiring significantly less data than base LLMs, making it ideal for data-limited applications.
Introduced TxGemma-Chat, a conversational model providing natural language reasoning and mechanistic explanations for therapeutic predictions.
Developed Agentic-Tx, an advanced therapeutic agentic system powered by Gemini 2.0, surpassing prior models on chemistry and biology reasoning benchmarks, including Humanity’s Last Exam and ChemBench.
By combining predictive accuracy with explainability and interactive reasoning, TxGemma advances AI-driven therapeutics, accelerating drug discovery and development.
ProteinDT: A Text-Guided AI Framework for Protein Design
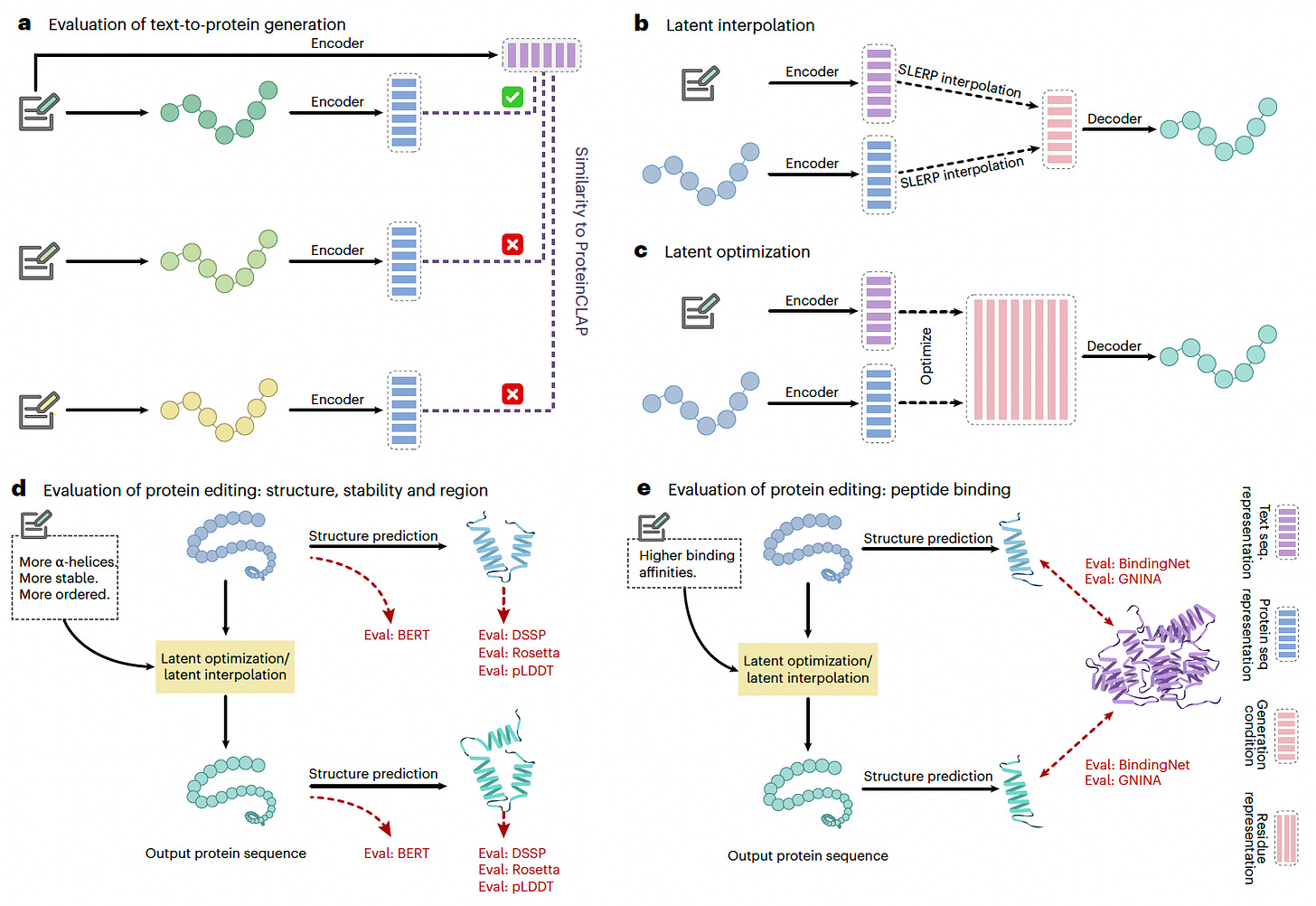
A new study introduces ProteinDT, a multimodal AI framework that integrates textual descriptions with protein sequences to enhance protein design. By leveraging contrastive learning and generative modeling, ProteinDT enables text-guided protein generation, editing, and property prediction, addressing a critical gap in AI-assisted protein engineering.
ProteinDT achieves over 90% accuracy in text-guided protein generation, demonstrating strong generalization across multiple protein design tasks.
Trained on SwissProtCLAP, a dataset of 441,000 text-protein pairs extracted from UniProt, aligning textual and protein sequence representations.
Utilized contrastive pretraining (ProteinCLAP) to map protein descriptions to structured protein representations, enhancing cross-modal understanding.
Applied generative models (autoregressive and diffusion-based) for conditional protein sequence generation from text inputs, achieving state-of-the-art retrieval accuracy.
Enabled zero-shot protein editing, outperforming baselines on 12 text-guided protein modification tasks, including secondary structure, stability, and peptide-binding adjustments.
Improved protein property prediction across six benchmark tasks, surpassing existing sequence-based models in secondary structure and evolutionary feature prediction.
By integrating textual knowledge with protein sequences, ProteinDT paves the way for AI-driven, text-guided protein engineering, accelerating advancements in synthetic biology and drug discovery.
DYNA: A Disease-Specific Language Model for Genomic Variant Interpretation
DYNA is a disease-specific fine-tuning framework that enhances genomic foundation models for variant effect prediction (VEP) in clinical genetics. By leveraging a Siamese neural network, DYNA fine-tunes protein and DNA language models on small disease-specific datasets, improving the classification of pathogenic versus benign genetic variants.

DYNA significantly outperforms existing VEP models in rare cardiovascular and RNA splicing variants, demonstrating superior disease-specific prediction accuracy.
Fine-tunes genomic foundation models with disease-specific data, enhancing their ability to distinguish between pathogenic and benign variants.
Outperforms baseline models in classifying rare missense variants for cardiomyopathies and arrhythmias, achieving significant performance gains in ClinVar datasets.
Uses a contrastive loss function for non-coding variants and a pseudo-log-likelihood ratio (PLLR) for protein variants, improving variant effect predictions.
Demonstrates strong generalization to unseen disease-relevant genes, expanding clinical applicability beyond pre-annotated datasets.
Enhances variant classification in regulatory genomics, particularly in splicing-related diseases, refining clinical decision-making.
By integrating disease-specific modeling with genomic language models, DYNA provides a scalable AI-driven approach for clinical variant interpretation and precision medicine.
Med3DVLM: A Vision-Language Model for 3D Medical Imaging
While previous VLMs focused on 2D data or struggled with the high computational cost of 3D inputs, Med3DVLM introduces innovations in architecture and training to overcome these hurdles.
Med3DVLM is a vision-language model tailored for 3D medical image analysis tasks like report generation and visual question answering.
It outperforms the current state-of-the-art across multiple benchmarks, including image-text retrieval (R@1: 61.00%), radiology report generation (METEOR: 36.42%), and open-ended VQA (METEOR: 36.76%).
Used a decomposed 3D convolutional encoder (DCFormer) to efficiently capture spatial features across large volumetric data, enabling high-resolution analysis at lower computational cost.
Replaced CLIP’s softmax-based contrastive loss with SigLIP, a sigmoid-based pairwise loss function that improved image-text alignment without requiring large negative batches, enhancing training stability on smaller medical datasets.
Integrated a dual-stream MLP-Mixer projector that fused low-level and high-level image features with ClinicalBERT-derived text embeddings, improving semantic alignment for tasks like VQA and report generation.
Achieved state-of-the-art performance on the M3D dataset, with substantial gains over M3D-LaMed in retrieval (R@1: 61.00% vs. 19.10%) and report generation (METEOR: 36.42% vs. 14.38%), while maintaining competitive BERTScores, indicating strong linguistic quality and content relevance.
Demonstrated superior accuracy in both open-ended and closed-ended VQA, especially in complex reasoning tasks such as identifying organ abnormalities and CT scan phases, showing its capacity for clinically meaningful interpretations.
Med3DVLM sets a new benchmark for scalable, multimodal AI in radiology by bridging the gap between 3D imaging and language, with clear implications for diagnostic support, education, and clinical efficiency.
iKraph: A Large-Scale Biomedical Knowledge Graph for AI-Powered Research
Unlike previous methods, which struggled with accuracy and comprehensiveness, iKraph uses an award-winning information extraction pipeline to transform PubMed abstracts into a structured KG with human-level precision. iKraph is a large-scale biomedical knowledge graph (KG) designed to integrate and structure vast amounts of biomedical literature and genomic data. This resource has already demonstrated its utility in drug repurposing, identifying COVID-19 treatments in real-time with substantial validation from clinical trials and publications.
Extracted over 30 million unique relations from more than 34 million PubMed abstracts, surpassing manually curated public databases in both scale and accuracy.
Integrated relation data from 40 public biomedical databases and high-throughput genomic datasets to enhance coverage and improve automated knowledge discovery.
Developed a probabilistic semantic reasoning (PSR) algorithm to infer indirect causal relationships between biomedical entities, enabling new hypothesis generation.
Conducted real-time drug repurposing for COVID-19 between March 2020 and May 2023, identifying approximately 1,200 candidate drugs within four months, with one-third validated by clinical trials or later PubMed publications.
Created a cloud-based platform for academic researchers, allowing efficient access to structured biomedical knowledge and facilitating AI-driven discoveries.
By structuring biomedical knowledge at an unprecedented scale, iKraph enables more effective information retrieval, automated hypothesis generation, and accelerated biomedical research.
AbMAP: A Transfer Learning Framework for Modeling Antibody Hypervariability
A new study introduces AbMAP, a transfer learning framework that adapts protein language models (PLMs) to better capture the hypervariable regions of antibodies. Unlike foundational PLMs, which struggle with modeling these regions due to their lack of evolutionary conservation, AbMAP focuses on refining CDR-specific representations through contrastive augmentation and multitask supervision.
AbMAP achieves state-of-the-art performance across tasks like antigen binding prediction, paratope identification, and antibody optimization, offering a scalable and data-efficient tool for antibody design.
Leveraged contrastive augmentation by performing in silico mutagenesis on complementarity-determining regions (CDRs), enhancing sensitivity to sequence variation while preserving structural relevance.
Trained a multitask Siamese transformer using antibody structural similarity and antigen binding profiles to create embeddings that encode both 3D structure and functional specificity.
Demonstrated a hit rate of 82% in optimizing SARS-CoV-2-binding antibodies, achieving up to 22-fold increases in binding affinity through iterative design and experimental validation.
Enabled accurate ΔΔG prediction for antibody variants with as little as 0.5% training data, outperforming foundational PLMs and one-hot baselines in low-data settings.
Revealed structure–function convergence across human B-cell repertoires, showing that individual repertoires, despite high sequence diversity, cluster in similar functional embedding space.
By fine-tuning general-purpose PLMs for antibody-specific challenges, AbMAP paves the way for more efficient therapeutic antibody design and deeper insights into immune system diversity.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun







Hi Luke,
I’ve been following your work at the intersection of AI and biomedical research, and I really admire how you break down complex advancements in health intelligence. Your recent discussions on AI-driven drug discovery, cognitive decline detection, and multimodal medical models are incredibly insightful.
I’m part of a team developing an AI-powered tool that streamlines literature review for biomedical research. It leverages the PubMed database to automatically retrieve, filter, and synthesize scientific papers into structured reports—including background, methodologies, key findings, and conclusions.
We’re currently in the beta phase and would love to invite experts like you to test our tool. If you're interested, send me 1-2 research keywords relevant to your work, along with your email, and I’d be happy to generate a customized literature review report for you—completely free.
You can read more about our project here: https://harryblackwood.substack.com/p/pubmed-ai-join-our-beta-testing-program?r=4tsyq1.
Looking forward to your thoughts!
Best regards