DeepMind's AlphaGenome, Microsoft AI's MAI-DxO, Arc Institute's State 🚀
Health Intelligence (HINT)
2025-06-30
🚀
AlphaGenome: A Unified DNA Sequence Model for Regulatory Variant Prediction
Google DeepMind with another banger. They present AlphaGenome, a deep learning model that predicts thousands of functional genomic tracks from 1 megabase of DNA sequence at single base-pair resolution. By bridging modalities such as gene expression, chromatin accessibility, splicing, and 3D genome architecture, AlphaGenome enables comprehensive prediction of variant effects across diverse regulatory mechanisms.
Trained on human and mouse genomes, AlphaGenome outperforms existing models on 24 out of 26 benchmarks for variant effect prediction, offering a more complete interpretation of non-coding variants that underlie many genetic diseases.
Predicted 5,930 human and 1,128 mouse genome tracks spanning 11 modalities, including RNA expression, detailed splicing features, histone modifications, transcription factor binding, and chromatin contacts, using a 1 Mb DNA context that captures distal regulatory elements.
Employed a U-Net-inspired architecture combining convolutional layers for local sequence patterns and transformers for long-range dependencies, achieving single-base resolution while modeling interactions up to 1 Mb away.
Demonstrated state-of-the-art performance in splicing variant effect prediction by simultaneously modeling splice site presence, usage, and junctions, enabling accurate detection of disease-causing splicing disruptions in empirical cases from GTEx and ClinVar datasets.
Achieved superior prediction of gene expression effects for eQTLs, enhancer-gene interactions, and polyadenylation QTLs compared to leading models like Borzoi, significantly improving the ability to assign regulatory mechanisms to GWAS variants, especially for rare and distal non-coding variants.
AlphaGenome sets a new benchmark for interpreting the regulatory code of the genome, providing researchers with a unified, multimodal tool for predicting the molecular impact of non-coding genetic variation.
State: A Multi-Scale AI Model for Predicting Cellular Responses to Perturbations
A new study introduces State, an AI architecture designed to predict cellular responses to genetic, chemical, and signaling perturbations with unprecedented accuracy across diverse biological contexts. Trained on data from over 100 million perturbed cells and 167 million observational single-cell profiles, State addresses key challenges of cellular heterogeneity and experimental noise that have limited prior models’ ability to generalize.
State combines two modules, a State Transition model (ST) leveraging transformer-based self-attention to model perturbation effects on sets of cells, and a State Embedding model (SE) learning robust cell representations from large-scale observational data, to create a flexible, scalable virtual cell model.
Achieved consistent performance improvements over baselines in predicting gene expression changes and differentially expressed genes across datasets of drug, cytokine, and genetic perturbations, increasing perturbation discrimination scores by 54% and precision-recall metrics by up to 184%.
Demonstrated strong zero-shot generalization capabilities, using its cell embeddings to predict perturbation effects in new cellular contexts without requiring perturbation data from those contexts, ranking perturbations by effect size with over 17% improvement in Spearman correlation across multiple datasets.
Detected cell type-specific responses to perturbations, accurately identifying differentially expressed genes unique to held-out cell lines and predicting context-specific effects of clinically relevant drugs such as Trametinib in BRAF-mutant melanoma cells.
Provided a comprehensive evaluation framework, Cell-Eval, using biologically relevant metrics to benchmark performance across key outputs of perturbation experiments, including gene expression counts, differential expression statistics, and perturbation effect magnitudes.
By effectively modeling both known and hidden sources of cellular heterogeneity, State advances the development of virtual cell models capable of guiding personalized treatment predictions and accelerating drug discovery through scalable, in silico simulations of cellular perturbations
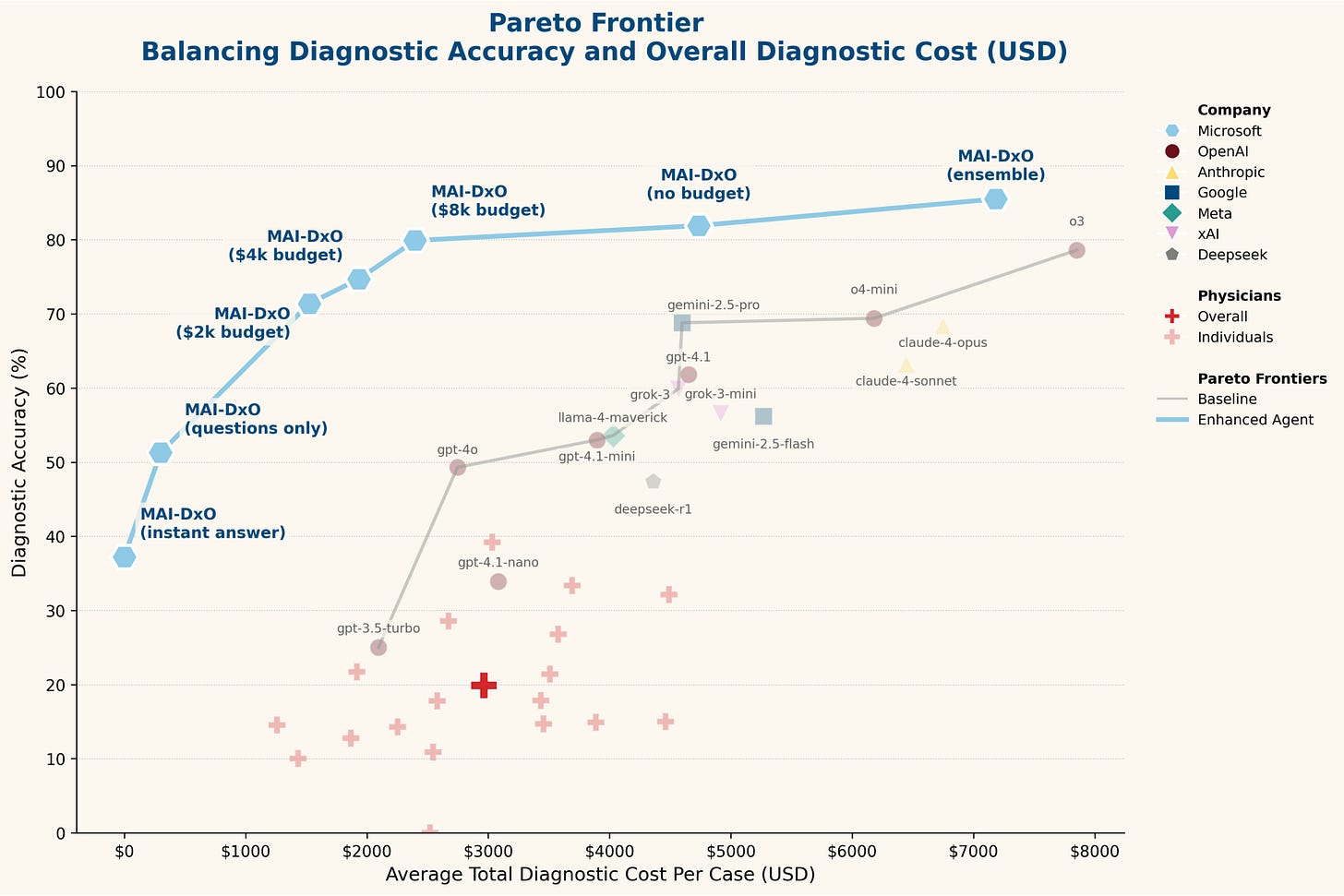
Sequential Diagnosis with Language Models
Microsoft AI introduces the Sequential Diagnosis Benchmark (SDBench), an interactive framework that turns 304 challenging New England Journal of Medicine CPC cases into stepwise diagnostic encounters. Instead of static vignettes, SDBench requires AI or physicians to iteratively ask questions, order tests, and decide when to commit to a diagnosis, reflecting real-world diagnostic reasoning.
Alongside this benchmark, the MAI Diagnostic Orchestrator (MAI-DxO) was developed to emulate a virtual panel of doctors, significantly improving diagnostic accuracy and cost efficiency compared to both practicing physicians and standard AI models.
Converted NEJM CPC cases into sequential simulations where diagnostic agents must actively choose questions and tests, capturing the complexity of iterative clinical reasoning often lost in traditional multiple-choice evaluations.
Demonstrated that practicing physicians averaged only 20% diagnostic accuracy at a cost of $2,963 per case, while off-the-shelf language models varied widely, with the strongest (o3) reaching 78.6% accuracy but incurring $7,850 per case.
Developed MAI-DxO, an orchestration system simulating specialized virtual doctors who maintain differentials, challenge biases, and enforce cost-conscious strategies, achieving 85.5% accuracy with ensemble techniques while reducing average diagnostic costs by over 20% compared to off-the-shelf AI.
Validated performance improvements across diverse models (OpenAI, Gemini, Claude, Grok, DeepSeek, Llama) and confirmed gains on a held-out test set of recent NEJM cases, demonstrating that structured reasoning reduces premature closure and improves both accuracy and cost-effectiveness.
By bridging the gap between static benchmarks and dynamic clinical practice, SDBench and MAI-DxO offer a powerful foundation for developing AI systems capable of safe, efficient, and clinically realistic diagnostic reasoning.
SHEPHERD: Few-Shot Learning for Rare Disease Diagnosis
A recent study presents SHEPHERD, an innovative few-shot deep learning model designed to aid the diagnosis of rare genetic diseases, which collectively affect hundreds of millions but suffer from chronic underdiagnosis.
SHEPHERD integrates patient phenotypes with a knowledge graph of genes and diseases, enabling accurate diagnosis even when labeled patient data are scarce. By learning directly from phenotype terms and simulated rare disease patients, SHEPHERD demonstrates unprecedented performance across challenging diagnostic tasks.
Trained on a synthetic cohort of over 40,000 simulated rare disease patients representing more than 2,000 rare diseases, enabling robust learning despite limited real-world data availability.
Leveraged a knowledge-guided graph neural network architecture to embed patient phenotypes within a knowledge graph, aligning patients near their causal genes and similar cases in a shared latent space.
Achieved top-rank causal gene identification in 40% of patients from the Undiagnosed Diseases Network, and placed the correct gene among the top five predictions for 77.8% of patients with atypical or novel disease presentations.
Outperformed existing state-of-the-art algorithms and large language models across multiple benchmarks, reducing the number of genes experts must evaluate by up to 23% and effectively retrieving "patients-like-me" for case comparison and validation.
SHEPHERD offers a transformative AI-driven tool for shortening the diagnostic odyssey in rare diseases, bridging knowledge gaps through few-shot learning and scalable simulation-based training.
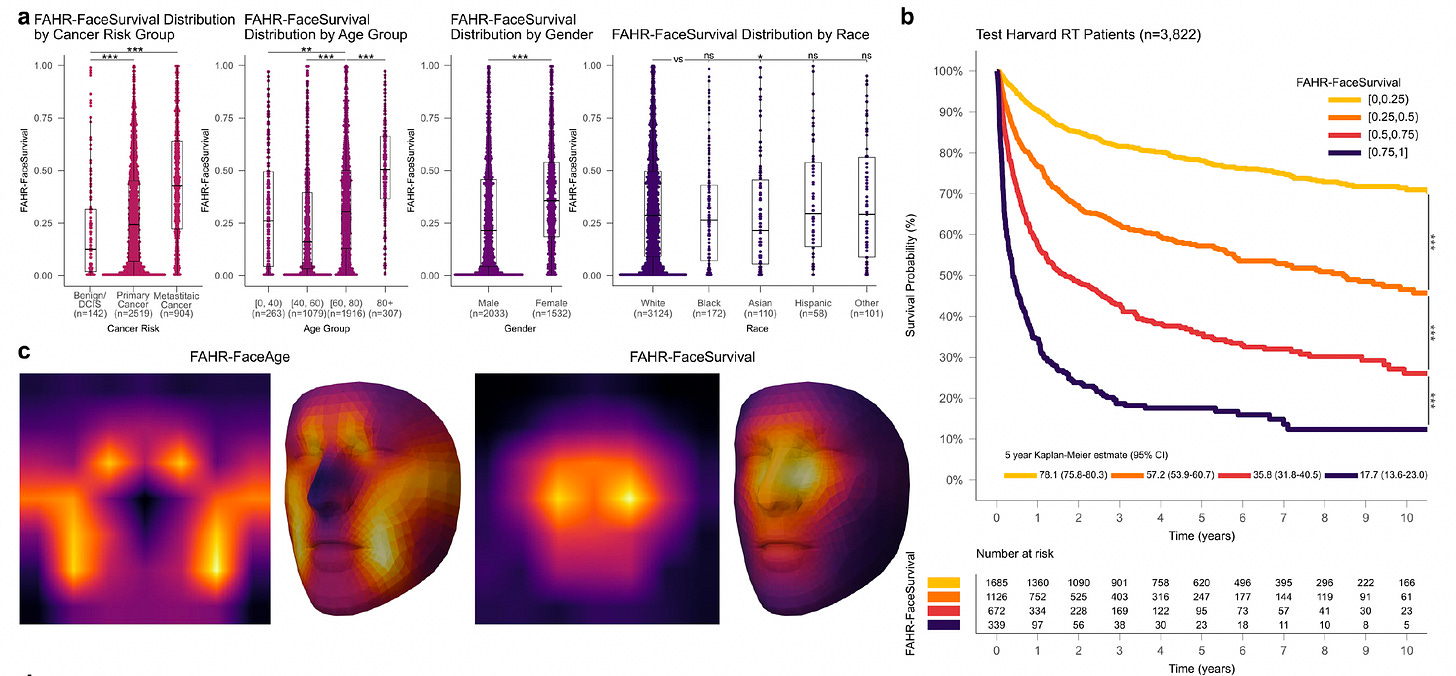
FAHR-Face: Foundation AI for Health Recognition from Facial Photographs
Researchers developed FAHR-Face, a foundation model trained on over 40 million facial images, enabling accurate estimation of biological age and prediction of mortality risk from everyday facial photographs.
By fine-tuning the model on age-labeled public datasets and photos of cancer patients, they created FAHR-FaceAge for estimating biological aging and FAHR-FaceSurvival for predicting survival outcomes. These models demonstrated robust, generalizable performance across age, sex, race, and cancer subgroups, offering scalable, non-invasive health biomarkers.
Trained a masked autoencoder on 40 million facial images, leveraging self-supervised learning to capture nuanced facial features related to aging and disease risk without requiring large, labeled clinical datasets.
Fine-tuned FAHR-FaceAge on nearly 750,000 public images, achieving a mean absolute error of 5.1 years in age estimation—outperforming previous models—and validated it on 38,211 cancer patients, where deviations between predicted and chronological age correlated significantly with survival outcomes.
Developed FAHR-FaceSurvival by fine-tuning on over 34,000 cancer patient photos, finding that patients in the highest risk quartile had more than three times the mortality of those in the lowest quartile, with strong performance validated in an independent cohort of nearly 5,000 patients.
Demonstrated that FAHR-FaceAge and FAHR-FaceSurvival provided distinct, complementary prognostic information from different facial regions, with low correlation and minimal feature overlap, enabling improved survival prediction when combined in multivariable models.
FAHR-Face highlights the potential of foundation models trained on consumer-grade facial photos to generate inexpensive, scalable biomarkers of biological age and mortality risk, paving the way for accessible health monitoring and early risk stratification using a single facial image.
MedCite: Enabling Verifiable Medical QA with LLM-Generated Citations
A new study introduces MedCite, the first end-to-end system for large language models (LLMs) to generate verifiable answers with citations for medical question-answering (QA). Unlike current medical LLMs that provide answers without evidence, MedCite combines retrieval-augmented generation and novel citation-seeking strategies to ensure that each statement in an answer is supported by relevant medical literature, improving trustworthiness and reliability.
Extensive evaluations on BioASQ and PubMedQA datasets show that MedCite significantly outperforms existing citation methods in precision and recall of citations, without compromising answer correctness. By leveraging a multi-pass citation approach and a hierarchical retriever strategy, MedCite aligns generated statements with accurate citations from trusted sources like PubMed.
Developed a multi-pass citation generation process that combined both parametric (LLM-based) and non-parametric (retrieval-based) citations, leading to up to 47.39% improvement in recall and 31.61% in precision compared to baselines.
Demonstrated that integrating retrieval-augmented generation (RAG) with post-generation citation seeking improved both answer correctness and citation quality, achieving accuracies up to 94.34% when ground-truth documents were available.
Evaluated different citation-seeking strategies, finding that re-retrieval followed by LLM reranking of documents produced the highest citation precision (up to 60.95%), highlighting the importance of carefully designed retrieval pipelines.
Validated automatic citation evaluation methods by correlating them with expert annotations from medical doctors, confirming that MedCite’s evaluation framework reliably reflects domain-expert judgments.
By enabling LLMs to generate verifiable, evidence-backed answers, MedCite sets a new standard for trustworthy AI systems in medicine, paving the way for safer clinical decision support and improved patient care.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun