CellVoyager, Interpolation for Restoring 3D Images, Efficient Foundational Model for Single-Cell Omics 🚀
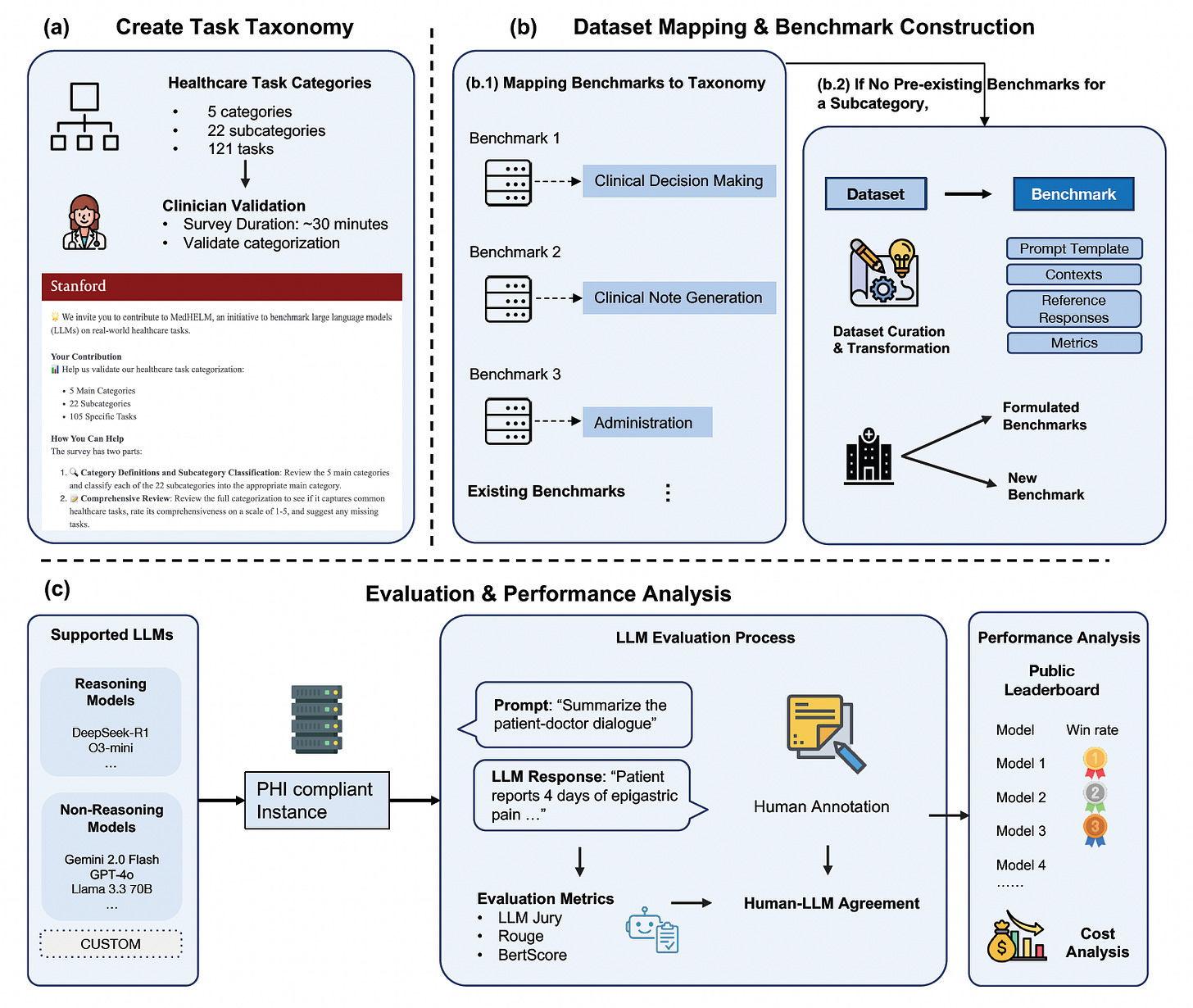
Health Intelligence (HINT)
2025-06-09
🚀
CellVoyager: An Autonomous AI Agent for Single-Cell Discovery
Stanford research introduces CellVoyager, an AI agent that autonomously explores single-cell RNA-seq (scRNA-seq) datasets to generate novel biological insights. Unlike prior agents that require user prompts, CellVoyager takes in processed data and a record of past analyses to independently design, execute, and refine new hypotheses within a Jupyter notebook environment.
Evaluated on a benchmark of 50 studies and three peer-reviewed case studies, CellVoyager consistently generated creative, biologically relevant findings not explored in the original papers.
Outperformed GPT-4o and o3-mini by up to 20% on the CellBench benchmark, predicting which analyses authors would conduct using only the biological background of a study.
Generated novel findings in expert-reviewed case studies, such as increased pyroptosis priming in CD8+ T cells during COVID-19 and transcriptional noise elevation with age in the brain’s subventricular zone.
Incorporated prior analyses to avoid redundancy, using vision-language models to interpret results and iteratively refine exploration blueprints during hypothesis testing.
Enabled human-in-the-loop interaction, successfully improving analyses based on expert feedback, demonstrating collaborative potential in biomedical research.
By autonomously navigating the vast analytical space of single-cell datasets, CellVoyager offers a scalable solution for uncovering hidden insights in both new and existing biological data.
BactoChat: A Foundation Model for Bacterial Gene Function Prediction
Researchers introduce BactoChat, a protein language model tailored to bacterial proteins, pre-trained on over 46 million bacterial sequences across 40,000 species. By capturing bacterial-specific evolutionary and structural patterns, BactoChat surpasses generalist models in functional prediction for bacterial genes, especially those with no known annotations.
BactoChat is a foundation model focused exclusively on bacteria, addressing the annotation gap in the microbial protein universe with high interpretability and generalizability.
Pre-trained on UniRef90 bacterial clusters using a 1.2 billion parameter transformer, yielding embeddings that better capture structure-function relationships in bacterial proteins.
Outperformed generalist models like ESM-1b and ProtBERT in protein function prediction, improving F1 score by 15% and demonstrating better zero-shot performance on unannotated genes.
Produced meaningful attention maps and embeddings that corresponded with conserved protein domains, enabling interpretable predictions and functional discovery.
Enabled new biological insights, such as identifying the sporulation regulator SpoIIID in previously unannotated Firmicutes genes, confirmed through conserved motif and domain analysis.
By specializing in the bacterial proteome, BactoChat provides a powerful tool for understanding microbial biology, with applications in microbiome research, antimicrobial resistance, and synthetic biology.
xTrimoGene: An Efficient and Scalable Foundation Model for Single-Cell Omics
The study presents xTrimoGene, a general-purpose foundation model designed for scalable and accurate representation learning across single-cell omics. Trained on over 10 million scRNA-seq profiles using a novel asymmetric encoder-decoder architecture, xTrimoGene addresses the high sparsity and dimensionality of single-cell data. It supports multiple downstream tasks across tissues and species while reducing computational cost.
xTrimoGene achieves state-of-the-art performance across eight benchmarks, including cell type annotation, perturbation prediction, and spatial transcriptomics alignment.
Adopted an asymmetric encoder-decoder structure, compressing input from 20,000 to 1,200 dimensions and reducing training FLOPs by 70% compared to transformer baselines.
Leveraged a denoising pretraining objective, learning to reconstruct masked gene expression values from sparse inputs, improving generalization to unseen cell types and datasets.
Outperformed prior methods on human and mouse datasets for tissue-level classification, perturb-seq outcome prediction, and cross-modality matching, with consistent accuracy gains.
Enabled high-throughput, memory-efficient inference with a 16x speedup, making it suitable for deployment at biobank scale and integration with other omics modalities.
xTrimoGene provides a fast and generalizable solution for large-scale single-cell analysis, unlocking the potential of foundation models for diverse biological discovery tasks.
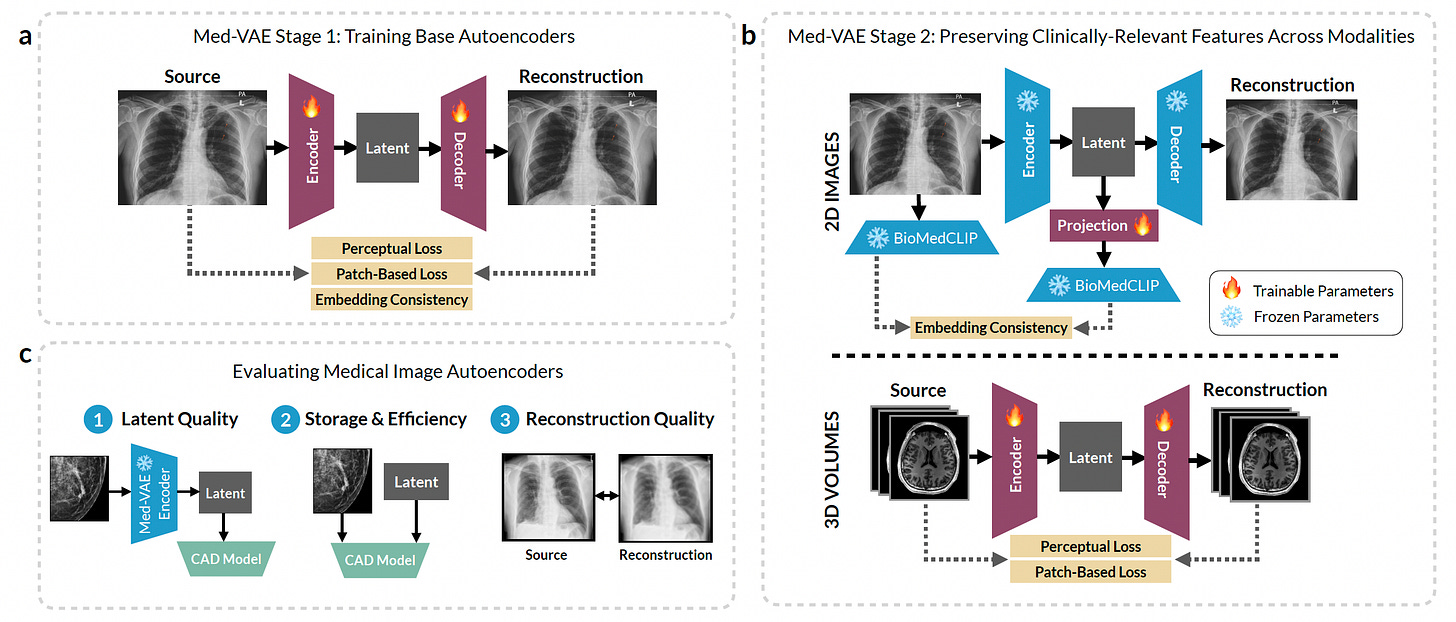
InterpolAI: Deep Learning-Based Interpolation for Restoring 3D Biomedical Images
A new study presents InterpolAI, an optical flow-based AI model that restores missing or damaged biomedical image slices to improve 3D tissue reconstruction. The model leverages large-motion frame interpolation adapted from video processing to synthesize realistic intermediate images between two undamaged slices.
InterpolAI significantly improves spatial continuity and cellular detail across a variety of imaging modalities, organs, and species.
Restored missing histology, IHC, light-sheet, ssTEM, and MRI slices with superior accuracy to linear and XVFI interpolation methods, preserving critical microanatomical features like ducts, blood vessels, and cell nuclei.
Reduced interpolation error across skipped slides by over 50% compared to other methods, as quantified by 13 Haralick texture features and validated by cell count accuracy and Euclidean distances from authentic images.
Removed tissue damage artifacts and stitching inconsistencies, particularly in ssTEM and light-sheet microscopy, enabling clearer 3D reconstructions of structures such as brain synapses and pancreatic ducts.
Enabled realistic 3D visualizations of complex tissues by maintaining structural fidelity in interpolated volumes, improving accuracy for downstream spatial biology applications.
InterpolAI demonstrates how optical flow-inspired deep learning can fill in gaps in biomedical image stacks, enabling higher-fidelity reconstructions without requiring exhaustive physical imaging.
VesNet: Self-Supervised Learning of Vascular Topology from 3D Microscopy
Researchers introduce VesNet, a self-supervised learning framework for extracting detailed vascular topology from 3D biomedical images without manual annotations.
Built on a contrastive learning strategy, VesNet learns to recognize tubular structures like blood vessels by distinguishing between line-like and blob-like features. The method enables generalizable and scalable vessel segmentation across imaging modalities and species.
Learned vessel-relevant representations from unlabeled images using synthetic 3D training data that mimic line-based vascular structures and local image artifacts like branching and bifurcation.
Outperformed supervised and self-supervised baselines on 3D vessel segmentation tasks across multiple organs, species, and modalities, including mouse brain light-sheet microscopy and zebrafish multiphoton data.
Preserved fine vascular topology such as endpoints, bifurcations, and connectivity, enabling more accurate graph-based analysis compared to voxel-level metrics alone.
Maintained robustness when fine-tuned on small amounts of labeled data, enabling sample-efficient training and transfer to real-world biomedical datasets.
VesNet offers a scalable and annotation-efficient solution for 3D vascular mapping, enabling better understanding of microvascular networks in health and disease.
DeepIFC: Virtual Fluorescent Labeling of Blood Cells via Imaging Flow Cytometry
A new study presents DeepIFC, a deep learning framework that performs virtual fluorescent labeling of blood cells using imaging flow cytometry (IFC) data.
DeepIFC reconstructs fluorescent marker images from brightfield and darkfield channels, enabling marker prediction without physical staining. This approach facilitates label-free cell typing and reduces reliance on costly or destructive labeling procedures.
Reconstructed seven distinct fluorescent marker images—including CD45, CD14, and CD3—using only label-free brightfield and darkfield images as input.
Accurately identified major blood cell types such as T cells, monocytes, and B cells, achieving high correlation with traditional marker-based methods across three donors.
Demonstrated strong generalization by identifying rare or unseen cell types, including triple-negative cells, even when such types were not part of the training data.
Enabled interpretable feature extraction, with the model’s internal representations forming well-separated clusters in UMAP space corresponding to specific cell types.
DeepIFC introduces a powerful tool for virtual labeling in flow cytometry, reducing dependence on reagents while preserving high-throughput and cell-type specificity.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun