Brain-to-Voice Prosthesis, MRI version of Total Segmentator, 100B Protein Design Model 🚀
Health Intelligence (HINT)
2025-04-07
🚀
New Developments in Research
Streaming Brain-to-Voice Neuroprosthesis for Restoring Naturalistic Communication
I thought this was so cool. UC Berkeley and UCSF comes up with a brain-to-voice neuroprosthesis that enables low-latency, continuous speech synthesis directly from cortical activity in individuals with severe paralysis.
Using a 253-channel ECoG array and deep learning models, the system decodes neural signals into fluent speech and text outputs in real time.
This marks a significant advance in restoring naturalistic communication without requiring any vocalization during training or use.
Achieved streaming speech synthesis in 80-ms increments using a bimodal RNN transducer architecture, enabling simultaneous speech and text decoding aligned with the participant's silent speech attempts.
Delivered real-time speech at a median rate of 47.5 to 90.9 words per minute with significantly reduced latencies (as low as 1.01 seconds), outperforming previous models that required sentence-level buffering and delayed synthesis.
Generalized across modalities by successfully applying the decoder to single-unit intracortical data and surface electromyography, demonstrating its adaptability beyond ECoG-based systems.
Operated continuously with implicit speech detection, synthesizing speech only during volitional attempts while maintaining low false-positive rates, enabling long-form naturalistic use outside structured trials.
This neuroprosthesis represents a major step toward restoring spontaneous, real-time spoken interaction for individuals with speech loss due to neurological injury.
MESA: Ecology-Inspired Multiomics Framework for Spatial Tissue Analysis
A new study introduces MESA (Multiomics and Ecological Spatial Analysis), a computational framework that combines ecological metrics with spatial multiomics data to quantify cellular diversity across tissue states.
MESA enables high-resolution mapping of cellular neighborhoods by integrating spatial proteomics and transcriptomics, offering new insights into tissue remodeling and disease progression.
Designed as a Python package, MESA provides systematic tools to explore spatial patterns linked to functional outcomes like immune response and cancer subtype.
Adapted ecological biodiversity indices to quantify cellular diversity across spatial scales, introducing metrics such as multiscale diversity index (MDI), global diversity index (GDI), and diversity proximity index (DPI) to detect tissue remodeling in autoimmune disease, colorectal cancer, and liver cancer.
Integrated spatial-omics data with single-cell RNA-seq using MaxFuse to generate in silico multiomics profiles, allowing detection of distinct cellular neighborhoods and subniches missed by conventional clustering methods.
Revealed CRC subtype differences by identifying Treg–macrophage cohabitation patterns and macrophage transcriptional states within tumor diversity hot spots, significantly improving survival prediction beyond traditional pathologist annotations.
Demonstrated generalizability across tissue types and spatial omics platforms (CODEX, CosMx SMI), uncovering new ligand–receptor interactions in HCC and highlighting macrophage–T cell communication as a key axis of disease progression.
MESA bridges ecological theory and multiomics to systematically decode spatial tissue architectures, enhancing our ability to study complex cellular environments in health and disease.
RelCon: A Motion Foundation Model Using Relative Contrastive Learning for Wearables
Apple, UIUC, MIT researchers present RelCon, the first motion foundation model trained on 1 billion wearable accelerometry signals using a novel relative contrastive learning approach.
By introducing a learnable motif-based distance function and leveraging sensor-specific augmentations, RelCon captures temporal dynamics and semantic similarities in human movement.
The model demonstrates strong generalization across tasks like gait regression and human activity recognition using data from over 87,000 participants in the Apple Heart and Movement Study.
Trained a neural distance function to capture motion motif similarity with invariance to sensor orientation, using reversible normalization and sparse attention to enhance semantic precision in time-series matching.
Introduced a relative contrastive loss that ranks the similarity of all candidate sequences, avoiding hard binary labeling and enabling nuanced modeling of within- and between-subject temporal relationships.
Outperformed 11 state-of-the-art models across six diverse downstream datasets, achieving top results in stride velocity and double support time regression, as well as 16-class activity classification at both subsequence and workout levels.
Demonstrated superior generalizability in benchmarking evaluations, surpassing large-scale models pretrained on UK Biobank and achieving top F1 scores even when test sensors were positioned differently from training sensors.
RelCon sets a new standard for wearable motion analysis, offering a compact and scalable foundation model for diverse health and activity monitoring tasks.
xTrimoPGLM: A Unified 100B Parameter Foundation Model for Protein Understanding and Design
BioMap has a new study introduces xTrimoPGLM, a 100-billion-parameter protein language model trained on 1 trillion tokens using a unified framework that integrates masked language modeling and autoregressive generation.
Unlike prior models limited to either protein understanding or generation, xTrimoPGLM enables both tasks by combining bidirectional and unidirectional training objectives in a single architecture.
This model sets new benchmarks across 18 tasks and powers xT-Fold, a state-of-the-art structure prediction tool.
Achieved top performance in 15 of 18 protein understanding tasks spanning structure, function, interaction, and developability by jointly optimizing masked and generative objectives across pretraining phases.
Enabled high-accuracy and fast structure prediction with xT-Fold, surpassing ESMFold and OmegaFold on CAMEO and CASP15 benchmarks, while offering 4-bit quantization and FlashAttention for efficient inference.
Generated diverse and structurally novel protein sequences with low sequence identity and high TM-scores, outperforming ProGen2 in confidence, diversity, and structural alignment with natural proteins.
Fine-tuned sequences using supervised and reinforcement self-training (SFT and ReST) to align with specific biochemical properties like fluorescence, fitness, and temperature stability, outperforming existing models in targeted protein design.
xTrimoPGLM sets a new foundation for unified protein modeling, offering scalable solutions for both interpretation and synthesis of biologically meaningful sequences.
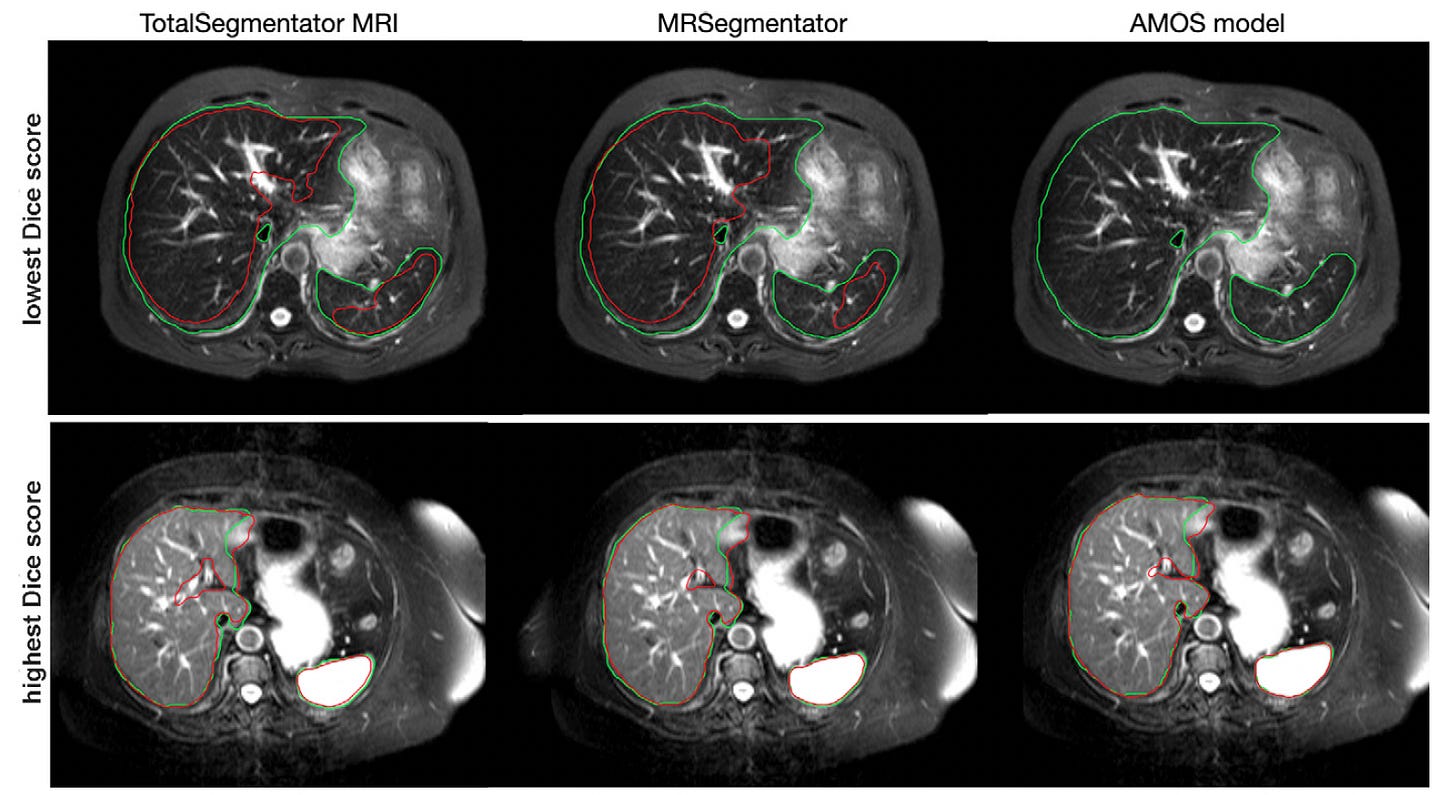
TotalSegmentator MRI: Robust, Sequence-Independent MRI Segmentation at Scale
After a successful TotalSegmentator CT, TotalSegmentator MRI is released as an open-source tool that automatically segments 80 major anatomic structures from MRI scans, regardless of imaging sequence.
Built on the nnU-Net framework and trained on 1,143 MRI and CT scans, the model achieves state-of-the-art performance across internal and external test sets.
This tool extends the capabilities of TotalSegmentator CT to MRI, enabling scalable, consistent, and clinically useful volumetric analysis.
Reached a Dice score of 0.839 on internal MRI test data and significantly outperformed existing models like MRSegmentator and AMOS across 40 and 13 shared structures, respectively, with P-values < .001 in both comparisons.
Maintained high accuracy across diverse datasets, achieving Dice scores up to 0.890 on external MRI benchmarks (CHAOS, AMOS), and showed strong generalizability across MRI sequences, scanner types, and body regions.
Demonstrated performance near parity with TotalSegmentator CT on CT data (Dice: 0.966 vs 0.970), while also supporting real-world MRI applications like opportunistic screening and age-related organ volume studies.
Enabled a large-scale analysis of abdominal organ aging patterns using 8,672 MRIs, uncovering statistically significant correlations between age and organ volume, including negative associations for liver and kidney volumes.
TotalSegmentator MRI offers a plug-and-play solution for automated MRI segmentation, supporting both research and clinical applications with reproducible and sequence-agnostic outputs.
MPAC: A Computational Framework for Inferring Pathway Activities from Multi-Omic Data
UW Madison researchers build MPAC, a pathway-based computational method that integrates multi-omic data to identify clinically meaningful patient subgroups.
Built upon and extending the PARADIGM algorithm, MPAC improves biological interpretability by filtering out spurious pathway signals and correlating pathway activity with survival and immune composition.
Applied to head and neck squamous cell carcinoma (HNSCC) data, MPAC uncovered an immune-response-related patient group not detectable from copy number or RNA data alone.
Improved pathway inference by integrating copy number and RNA-seq data through a refined pipeline that includes permutation filtering, GO enrichment, and downstream grouping of patients by pathway activity profiles.
Identified a subgroup of HPV+ HNSCC patients with activated immune response pathways, whose key proteins (e.g., CD28, CD86, FASLG) were associated with better overall survival and increased immune cell infiltration.
Validated these findings in an independent cohort of HPV+ patients, confirming that the immune-response subgroup and associated protein activities generalized beyond the training data.
Outperformed the original PARADIGM method by revealing pathway-based patient stratifications that PARADIGM could not detect, particularly those with prognostic and immunological significance.
MPAC offers a robust, interpretable tool for discovering biologically and clinically relevant patterns from multi-omic datasets, providing new opportunities for pathway-guided precision oncology.