2025-04-14
🚀
New Developments in Research
CodonTransformer: Context-Aware Codon Optimization Across Species
This is a new multispecies deep learning model that optimizes codon usage in DNA sequences for more efficient gene expression across a wide range of organisms.

Trained on over a million gene-protein pairs from 164 species, it leverages Transformer architecture to generate context-aware, host-specific DNA sequences with natural-like codon distributions. A user-friendly interface and customizable fine-tuning make CodonTransformer a powerful tool for synthetic biology and protein engineering.
Introduced a context-aware sequence encoding strategy combining amino acid, codon, and organism identity, enabling CodonTransformer to learn species-specific codon preferences from a large multispecies dataset.
Trained using a masked language modeling approach with an encoder-only BigBird architecture, allowing bidirectional optimization of DNA sequences for a given protein input.
Outperformed existing codon optimization tools (e.g., Genewiz, IDT, Twist, ICOR) in generating sequences with more natural codon usage patterns and fewer negative cis-regulatory elements across E. coli, yeast, plant, and human genomes.
Demonstrated predictive capabilities beyond sequence generation, showing significant correlation with ribosome stalling and protein fitness outcomes when evaluating synonymous mutations in E. coli.
CodonTransformer represents a scalable, open-access AI model for codon optimization, facilitating cross-species gene expression and advancing applications in synthetic biology, therapeutics, and protein design.
BoltzDesign1: Inverting Structure Prediction for Generalized Protein Binder Design
Researchers at MIT and EPFL introduced BoltzDesign1, a generative framework that repurposes the Boltz-1 structure prediction model to design protein binders across a wide array of molecular targets.
By optimizing the predicted distance distributions (distograms) rather than 3D atomic structures, BoltzDesign1 dramatically reduces computational costs while retaining high design quality. It supports binding to small molecules, nucleic acids, metal ions, and covalently modified proteins, expanding the landscape of biomolecular engineering.
Used only the Pairformer and Confidence modules from the Boltz-1 model to perform hallucination-based design, bypassing the need for diffusion model backpropagation while maintaining high accuracy.
Achieved higher in silico success rates and structural diversity than RfDiffusionAA when designing binders for four small-molecule targets (IAI, FAD, SAM, and OQO), with performance validated by AlphaFold3.
Demonstrated cross-model and self-consistency, and showed superior docking scores for several ligands compared to native binders, particularly when using interface-fixed designs optimized by LigandMPNN.
Extended design capabilities to complex targets such as B-DNA, metal ions (zinc and iron), and post-translational modifications, with designs forming specific interactions including hydrogen bonds and correct metal coordination geometries.
By leveraging a streamlined inversion of a structure prediction model, BoltzDesign1 establishes a scalable approach for designing diverse and functional protein binders across molecular classes.
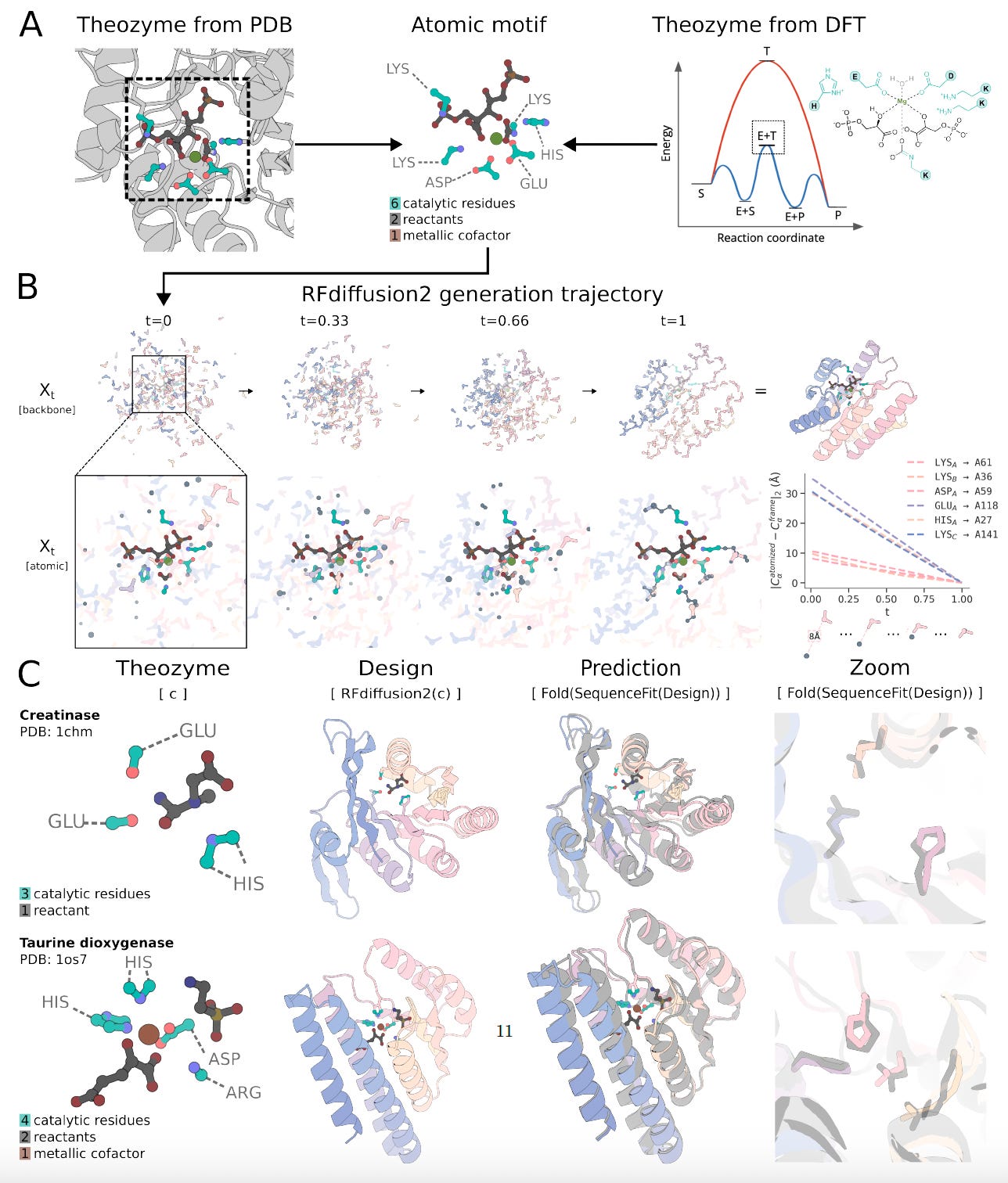
RFdiffusion2: Atomic-Level Protein Design for De Novo Enzyme Catalysis
Baker Lab unveils RFdiffusion2, a generative model that designs functional enzymes directly from atom-level active site descriptions.
Unlike previous methods that require residue-level inputs and sequence index specification, RFdiffusion2 scaffolds transition states without predefined residue positions or rotamer enumeration.
Trained on Protein Data Bank structures and evaluated with the new Atomic Motif Enzyme (AME) benchmark, RFdiffusion2 consistently generates novel scaffolds with catalytic activity across diverse reactions.
Enabled simultaneous inference of rotamers and sequence indices from atomic motifs, removing the need for inverse rotamer sampling or backbone motif indexing.
Outperformed prior methods by solving all 41 AME benchmark cases, compared to just 16 with RFdiffusion, and generated structurally novel scaffolds validated by Chai-1 structure prediction.
Demonstrated successful in vitro activity for four different reactions, including retroaldolases, cysteine hydrolases, and zinc-dependent hydrolases, with functional enzymes identified from fewer than 96 designs per case.
Incorporated novel conditioning features like relative solvent accessibility and partial ligand input, allowing precise control over ligand burial and conformation during structure generation.
RFdiffusion2 offers a powerful new approach to enzyme design by directly translating reaction mechanisms into active proteins, expanding the frontier of catalytic protein engineering.
MedSAM2: A Foundation Model for 3D Medical Image and Video Segmentation
A new study introduces a promptable segmentation foundation model designed for 3D medical images and videos, extending the Segment Anything framework to volumetric and dynamic medical data.
Fine-tuned on over 455,000 image-mask pairs and 76,000 video frames, it delivers superior performance across organs, lesions, and modalities. Integrated with a human-in-the-loop pipeline, MedSAM2 enables scalable and efficient medical annotation, cutting manual effort by more than 85%.
Adopted a memory-attentive architecture and hierarchical vision transformer encoder to process spatial and temporal information, enabling accurate segmentation of both 3D volumes and video sequences.
Outperformed SAM2.1 and EfficientMedSAM-Top1 in Dice similarity scores across CT, MRI, and PET scans, particularly for complex lesions and anatomies with heterogeneous appearances.
Enabled rapid annotation via iterative fine-tuning, reducing per-lesion segmentation time from 526 to 74 seconds in CT and from 520 to 65 seconds in MRI, and decreasing per-frame video annotation time from 102 to 8 seconds.
Deployed on platforms like 3D Slicer, JupyterLab, and Gradio, offering flexible local and cloud-based interfaces for integration into real-world clinical and research workflows.
By bridging the gap between generalist vision models and medical specificity, MedSAM2 offers a practical, open-source tool for high-throughput and high-accuracy segmentation across diverse imaging tasks.
SpatialAgent: An Autonomous AI Agent for Spatial Biology
Top companies and institutions teamed up to build SpatialAgent, a fully autonomous AI system designed to streamline spatial biology workflows.
Integrating large language models with dynamic tool execution, SpatialAgent handles experimental design, multimodal data analysis, and hypothesis generation.
Tested across diverse datasets, it matched or outperformed both human scientists and existing computational tools.
Outperformed human experts and leading methods in gene panel design, improving cell-type and spatial predictions by up to 19.1% and 47.1%, respectively, using adaptive reasoning and prebuilt plan templates.
Enhanced annotation of spatially resolved single cells and tissue niches in human heart samples by integrating MERFISH data, anatomical references, and multi-sample reasoning, achieving near-human accuracy while cutting cost and time.
Generated new biological insights from a DSS-induced colitis model by inferring cell-cell interactions, proposing fibroblast polarization via TGF-β and IL-11 signaling as key to tissue remodeling.
Improved experimental design for prostate cancer studies by selecting 100 genes that refined cell-type resolution and revealed laminin-integrin signaling pathways, enhancing tumor-immune interaction analysis.
SpatialAgent offers a scalable, interpretable, and collaborative platform for spatial biology, setting a new standard for autonomous discovery in biomedical research.
ATOMICA: A Universal Representation Model for Molecular Interactions
Harvard researchers present ATOMICA, a self-supervised geometric deep learning model that learns atomic-scale representations of molecular interfaces across diverse biomolecular modalities.
Trained on over two million interaction complexes, ATOMICA captures chemically meaningful interaction patterns and enables cross-modality generalization. This allows it to identify disease-relevant interaction modules and annotate uncharacterized binding sites in the dark proteome.
Modeled molecular complexes as hierarchical graphs that encode atomic and block-level features, enabling the model to represent interactions across proteins, small molecules, ions, nucleic acids, and lipids.
Trained using a denoising strategy that perturbs one molecule in a complex and learns to reconstruct the original configuration, allowing the model to internalize geometric and chemical principles of binding.
Constructed five modality-specific interaction networks (ATOMICANETs) and showed their utility in recovering disease-associated pathways in conditions like asthma, myeloid leukemia, and hypertrophic cardiomyopathy.
Annotated 2,646 previously uncharacterized binding sites in the dark proteome, including zinc finger motifs and transmembrane cytochrome domains, using fine-tuned models for ion and cofactor binding prediction.
ATOMICA provides a powerful, general-purpose tool for modeling molecular interfaces, revealing new biological insights and expanding our capacity to interpret unannotated regions of the proteome.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun