2025-06-16
🚀
Biomni: A General-Purpose Biomedical AI Agent
Stanford research introduces Biomni, a general-purpose biomedical AI agent that autonomously tackles a wide spectrum of research tasks across diverse subfields. It systematically mapped the biomedical action space by mining tens of thousands of publications across 25 domains to curate an environment with 150 specialized tools, 105 software packages, and 59 databases.
Integrating large language model reasoning with retrieval-augmented planning and code-based execution, Biomni composes dynamic workflows without predefined templates and achieves human-level zero-shot performance on benchmarks and real-world case studies.
Mapped the biomedical action space using an action discovery agent on thousands of papers, extracting and expert-curating essential tasks, tools, and databases.
Curated and validated an AI environment featuring 150 specialized biomedical tools, 105 preinstalled software packages, and 59 accessible databases via natural language queries.
Developed a generalist agent architecture that combined LLM-based tool selection, adaptive planning, and executable code composition for flexible, complex workflow execution.
Demonstrated human-level zero-shot performance across diverse Q&A benchmarks and autonomously executed end-to-end analyses—from wearable sensor data and multi-omics integration to wet-lab protocol design—in real-world case studies.
By uniting a comprehensive tool environment with adaptive agentic planning and code execution, Biomni paves the way for virtual AI biologists to augment human research productivity and accelerate biomedical discovery.
An artificial nervous system for communication between wearable and implantable therapeutics
Georgia Tech researchers introduce SWANS, a Smart Wireless Artificial Nervous System that harnesses the body’s ionic conduction pathways to link wearable sensors and implantable therapeutics into a coordinated network. By emitting controlled square-wave pulses (0–12 V) through stainless-steel microneedle patches, SWANS generates voltage gradients in tissue that selectively activate micrometer-scale transistor switches with over 15× greater power efficiency than Bluetooth Low Energy and NFC devices.
In vivo experiments in rats demonstrated precise, zero-template workflows to regulate dispersed implants across epidermal, subcutaneous, intraperitoneal, and gastrointestinal spaces—culminating in sensor-driven dual hind-leg motor control without detectable tissue damage .
Engineered a wearable hub with a flexible printed circuit board and microneedle interface that bypassed the stratum corneum to deliver programmable voltage pulses directly into subdermal tissue .
Reduced implant communication components to syringe-injectable footprints by using micrometer-scale NPN, PNP, MOSFET, and organic electrochemical transistor circuits, achieving near-zero passive current draw .
Demonstrated selective, multiplexed control over multiple implants by tuning pulse magnitude, width, and frequency to match distinct transistor gate thresholds, preventing cross-activation .
Validated in vivo in rats that SWANS could wirelessly trigger sciatic nerve stimulators for dual hind-leg motion and confirmed tissue health via histology after repeated 1 Hz pulses up to 10 ms at ≤ 10 V.
Can Large Language Models Match the Conclusions of Systematic Reviews?
A new study evaluates whether large language models (LLMs) can replicate expert-written conclusions from systematic reviews when given access to the same source studies. The authors created MedEvidence, a benchmark of 284 closed-form questions derived from 100 Cochrane systematic reviews spanning 10 medical specialties, each paired with the original studies and graded for evidence certainty.

By testing 24 LLMs of varying sizes, reasoning capabilities, and medical fine-tuning under zero-shot and expert-guided prompts, the study found that even leading models plateaued at around 60–62% accuracy and struggled with long contexts and uncertainty.
Constructed the MedEvidence benchmark by extracting question–answer pairs from Cochrane reviews (2014–2024) and linking each conclusion to its relevant studies, while capturing evidence quality via the GRADE framework.
Evaluated 24 LLMs (7B–671B parameters) using a two-step prompting approach that first solicited structured article summaries and then closed-form answers based on provided abstracts or full texts.
Observed that neither increased model size beyond 70B parameters nor explicit chain-of-thought reasoning reliably improved performance, and that medical fine-tuning often degraded accuracy.
Identified core failure modes: accuracy declined with longer input lengths, models displayed overconfidence and a lack of scientific skepticism toward low-quality evidence, and they underperformed on questions requiring uncertainty or recognizing insufficient data.
By exposing persistent gaps in LLMs’ abilities to critically appraise and synthesize clinical evidence, MedEvidence establishes a rigorous foundation for advancing AI-assisted systematic review automation.
MultiMorph: On-Demand Atlas Construction
A new study from MIT and MGH/HMS introduces MultiMorph, a machine learning method that constructs population-specific anatomical atlases rapidly and efficiently.
By replacing time-consuming iterative optimization with a single forward pass, MultiMorph enables researchers to generate high-quality atlases for any brain imaging dataset in seconds to minutes. This makes atlas construction accessible to researchers without requiring machine learning expertise or expensive computational infrastructure.
Employed a novel linear group-interaction layer within a UNet-based architecture to aggregate and share features across multiple input images, aligning them to a central, unbiased atlas space.
Integrated a centrality layer that explicitly removed global bias from estimated deformation fields, ensuring the generated atlas remained representative of the population group.
Leveraged synthetic neuroimaging data during training to generalize across diverse imaging modalities and unseen population groups at test time, including modalities not present during training.
Outperformed traditional and recent learning-based atlas construction methods by producing more accurate, unbiased atlases over 100 times faster, enabling on-the-fly generation for subgroup and conditional analyses.
MultiMorph democratizes high-quality atlas construction, paving the way for flexible, scalable studies of anatomical variability across populations and conditions.
Toward Scientific Reasoning in LLMs: Training from Expert Discussions via Reinforcement Learning
Princeton + Stanford researchers present an end-to-end approach to teach large language models (LLMs) scientific reasoning by training them on authentic expert discussions in genomics. The researchers introduce Genome-Bench, a benchmark derived from over a decade of real-world CRISPR forum threads, and apply reinforcement learning to enhance domain-specific problem solving.
Their framework shows that fine-tuning with structured expert dialogue significantly narrows the gap between open-source LLMs and human-level scientific reasoning.
Created Genome-Bench, a curated set of 3,332 multiple-choice questions covering experimental troubleshooting, tool use, and lab logistics, sourced from 11 years of CRISPR-related forum discussions.
Developed an automated pipeline to parse raw email threads, extract question-answer-context triplets, generate plausible distractors, and ensure high-quality, domain-accurate benchmarks for reinforcement learning.
Fine-tuned LLMs with a rule-based reward system that incentivized both answer correctness and explicit reasoning explanations, achieving over a 15% performance boost on Genome-Bench compared to base models.
Deployed an RL-trained router to dynamically select among multiple expert-tuned models, outperforming individual models and state-of-the-art commercial systems with an accuracy of 81.1% on Genome-Bench.
By grounding LLM training in real expert interactions, this study advances scientific reasoning capabilities in AI, offering a robust pathway toward domain-specialized and trustworthy language models.
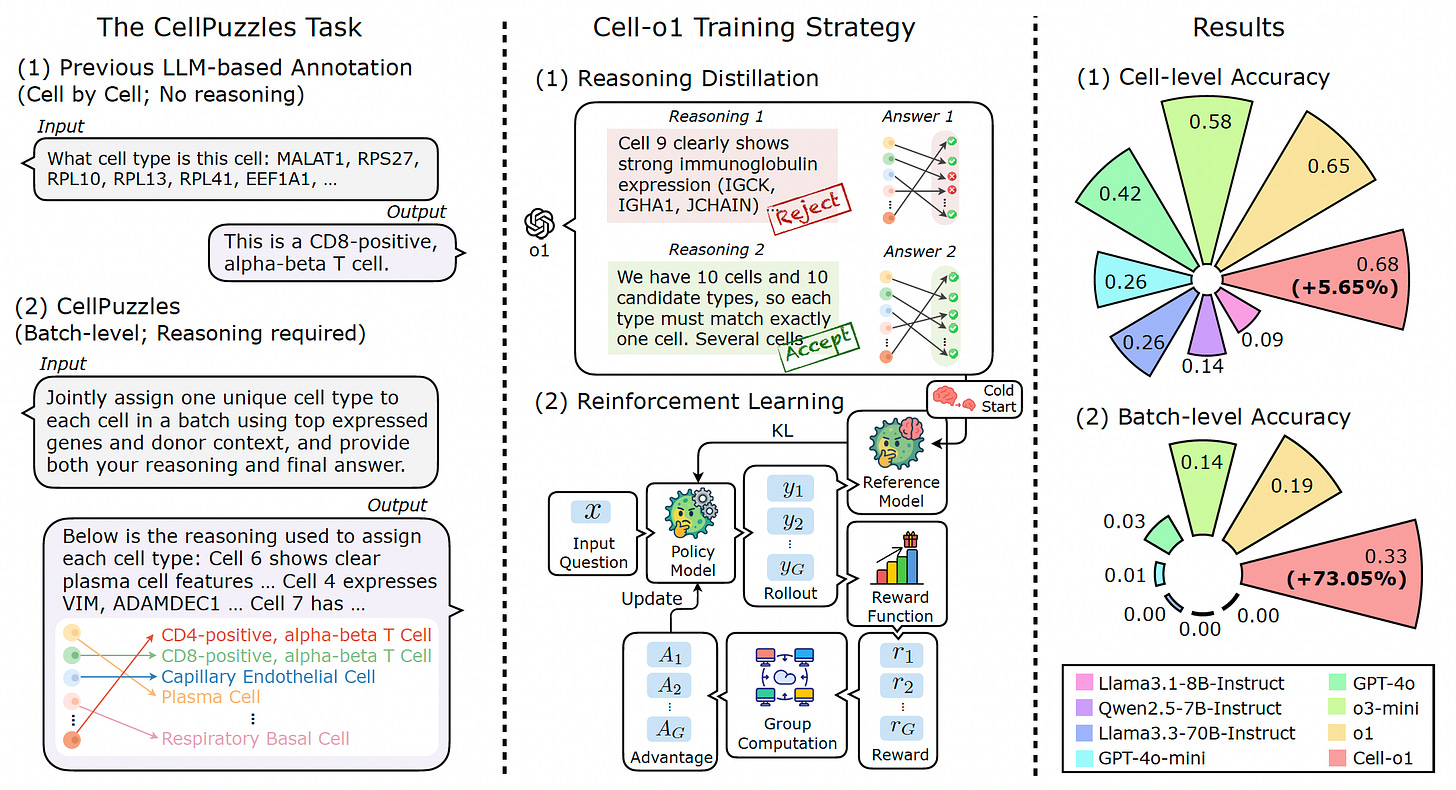
Cell-o1: Solving Single-Cell Reasoning Puzzles with Reinforcement Learning
A new study introduces Cell-o1, a reasoning-enhanced large language model designed to tackle the complex task of batch-level cell type annotation for single-cell RNA sequencing data.
Unlike conventional models that annotate cells independently and without explanation, Cell-o1 mimics expert workflows by jointly assigning unique labels to all cells in a batch and providing detailed reasoning traces. This approach sets a new state-of-the-art on the CellPuzzles benchmark, significantly improving both accuracy and interpretability in single-cell analysis.
Reformulated cell type annotation as the CellPuzzles task, requiring the model to reason across batch-level gene expression and context, ensuring each cell in a batch receives a unique, biologically consistent label.
Trained Cell-o1 through a two-stage process: first with supervised fine-tuning on high-quality reasoning traces distilled from a frontier LLM, then refined using reinforcement learning with batch-level rewards to promote consistency and expert-like annotation strategies.
Achieved state-of-the-art performance on both cell-level and batch-level accuracy metrics, outperforming leading closed-source and open-source LLMs and demonstrating robust generalization to unseen diseases such as melanoma and colorectal cancer.
Exhibited emergent expert behaviors like self-reflection and curriculum reasoning during annotation, allowing the model to revise intermediate conclusions and prioritize simpler assignments before tackling harder ones.
By bridging deep learning with expert reasoning, Cell-o1 enables accurate, interpretable, and scalable single-cell annotation, opening new avenues for trusted AI in biological discovery.
Love Health Intelligence (HINT)? Share it with your friends using this link: Health Intelligence.
Want to contact Health Intelligence (HINT)? Contact us today @ lukeyunmedia@gmail.com!
Thanks for reading, by Luke Yun